Read Table in R With Missing Values
Lesson 5. How to Address Missing Values in R
This lesson covers how to work with no information values in R.
Learning Objectives
At the end of this action, you lot will be able to:
- Understand why it is important to make note of missing information values.
- Be able to define what a
NAvalue is inRand how information technology is used in a vector.
What You lot Need
You lot need R and RStudio to consummate this tutorial. Also we recommend that you have an globe-analytics directory ready on your figurer with a /data directory within it.
- How to prepare R / RStudio
- Prepare your working directory
Missing Data - No Data Values
Sometimes, your information are missing values. Imagine a spreadsheet in Microsoft Excel with cells that are blank. If the cells are blank, you lot don't know for sure whether those data weren't collected, or someone forgot to fill them in. To account for data that are missing (non past mistake) you can put a value in those cells that represents no information.
The R programming language uses the value NA to represent missing data values.
planets <- c ( "Mercury" , "Venus" , "Earth" , "Mars" , "Jupiter" , "Saturn" , "Uranus" , "Neptune" , NA ) The default setting for well-nigh base functions that read information into R is to translate NA as a missing value.
Allow's have a closer expect at this using the boulder_precip data that y'all've used in the previous lessons. Please download the information again as in that location take been some changes made!
# download file download.file ( "https://ndownloader.figshare.com/files/9282364" , "data/boulder-precip-temp.csv" , method = "libcurl" ) Then y'all can open up the information.
# import data merely don't specify no information values - what happens? boulder_precip <- read.csv ( file = "data/boulder-precip-temp.csv" ) str ( boulder_precip ) ## 'information.frame': 18 obs. of four variables: ## $ ID : int 756 757 758 759 760 761 762 763 764 765 ... ## $ DATE : chr "8/21/thirteen" "8/26/13" "8/27/xiii" "9/ane/xiii" ... ## $ PRECIP: num 0.one 0.1 0.1 0 0.one one two.three 9.viii 1.ix i.four ... ## $ TEMP : int 55 25 NA -999 15 25 65 NA 95 -999 ... In the example below, notation how a hateful value is calculated differently depending upon on how NA values are treated when the data are imported.
# view mean values hateful ( boulder_precip $ PRECIP ) ## [i] 1.055556 hateful ( boulder_precip $ TEMP ) ## [ane] NA Notice that y'all are able to summate a mean value for PRECIP but TEMP returns a NA value. Why? Permit'south plot your data to figure out what might be going on.
library ( ggplot2 ) # are there values in the TEMP column of your data? boulder_precip $ TEMP ## [one] 55 25 NA -999 15 25 65 NA 95 -999 85 -999 85 85 ## [15] -999 57 lx 65 # plot the data with ggplot ggplot ( data = boulder_precip , aes ( x = Engagement , y = TEMP )) + geom_point () + labs ( title = "Temperature data for Boulder, CO" ) ## Alarm: Removed two rows containing missing values (geom_point). 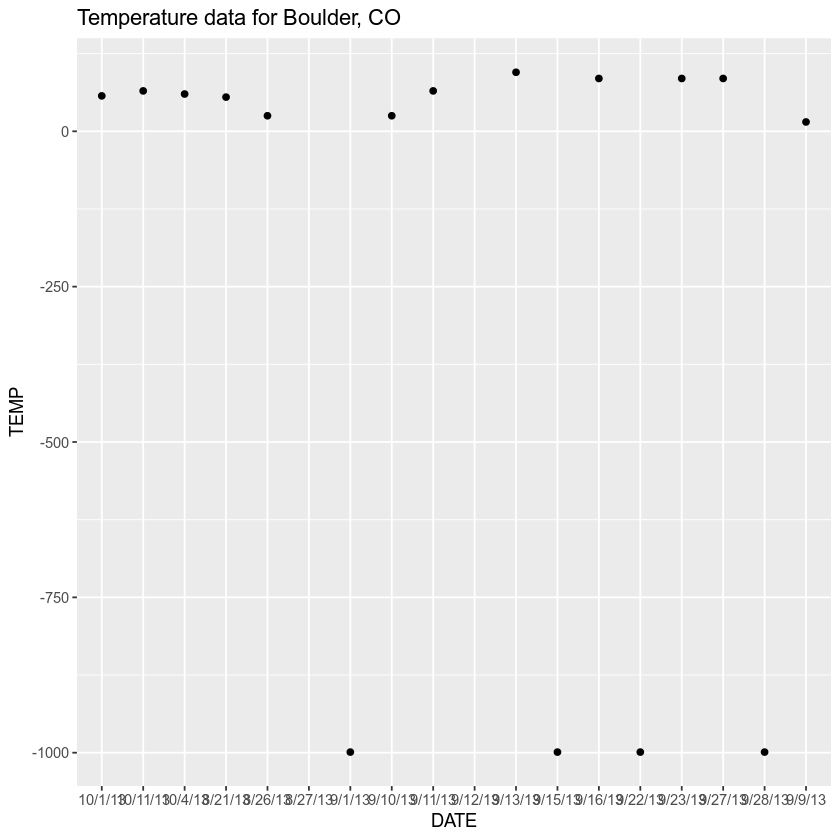
Looking at your data, it appears every bit if yous have some extremely big negative values hovering effectually -1000. However why did your hateful return NA?
When performing mathematical operations on numbers in R, most functions will return the value NA if the data you are working with include missing or nodata values.
Returning NA values allows yous to meet that you have missing information in your dataset. Y'all tin then decide how you want to handle the missing data. Youcan add the statement na.rm=Truthful to calculate the consequence while ignoring the missing values.
heights <- c ( 2 , iv , 4 , NA , vi ) mean ( heights ) ## [1] NA max ( heights ) ## [1] NA hateful ( heights , na.rm = TRUE ) ## [1] 4 max ( heights , na.rm = TRUE ) ## [i] 6 Let's endeavour to add together the na.rm argument to your code hateful calculation on the temperature column in a higher place.
# summate mean usign the na.rm argument mean ( boulder_precip $ PRECIP ) ## [1] 1.055556 mean ( boulder_precip $ TEMP , na.rm = TRUE ) ## [one] -204.9375 Data Tip: The functions, is.na(), na.omit(), and complete.cases() are all useful for figuring out if your data has assigned (NA) no-information values. Come across below for examples.
Then now you have successfully calculated the mean value of both atmospheric precipitation and temperature in your spreadsheet. However does the hateful temperature value (-204.9375 make sense looking at the data? It seems a bit low - you know that there aren't temperature values of -200 here in Boulder, Colorado!
Remembering the plot above you noticed that you had some values that were close to -1000. Looking at the summary below you meet the exact minimum value is -999.
# calculate mean usign the na.rm argument summary ( boulder_precip $ TEMP , na.rm = TRUE ) ## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's ## -999.0 -238.5 56.0 -204.ix 70.0 95.0 2 Finding & Assigning No Data Values
Sometimes, you'll find a dataset that uses another value for missing data. In some disciplines, for instance -999, is ofttimes used. If there are multiple types of missing values in your dataset, you tin can extend what R considers a missing value when it reads the file in using the "na.strings" argument.
Beneath use the na.strings argument on your data. Find that you can tell R that there are several potential ways that your information documents nodata values.
You lot can provide R with a vector of missing engagement values every bit follows:
c("NA", " ", "-999")
Thus R volition assign whatsoever calls with the values of cipher "", NA or -999 to NA. This should solve all of your missing data bug!
# import data but specify no data values - what happens? boulder_precip_na <- read.csv ( file = "information/boulder-precip-temp.csv" , na.strings = c ( "NA" , " " , "-999" )) boulder_precip_na $ TEMP ## [one] 55 25 NA NA 15 25 65 NA 95 NA 85 NA 85 85 NA 57 threescore 65 Does your new plot look meliorate?
# are there values in the TEMP column of your data? boulder_precip $ TEMP ## [1] 55 25 NA -999 15 25 65 NA 95 -999 85 -999 85 85 ## [xv] -999 57 lx 65 # plot the information with ggplot ggplot ( data = boulder_precip_na , aes ( x = Appointment , y = TEMP )) + geom_point () + labs ( title = "Temperature data for Bedrock, CO" , subtitle = "missing data accounted for" ) ## Warning: Removed 6 rows containing missing values (geom_point). 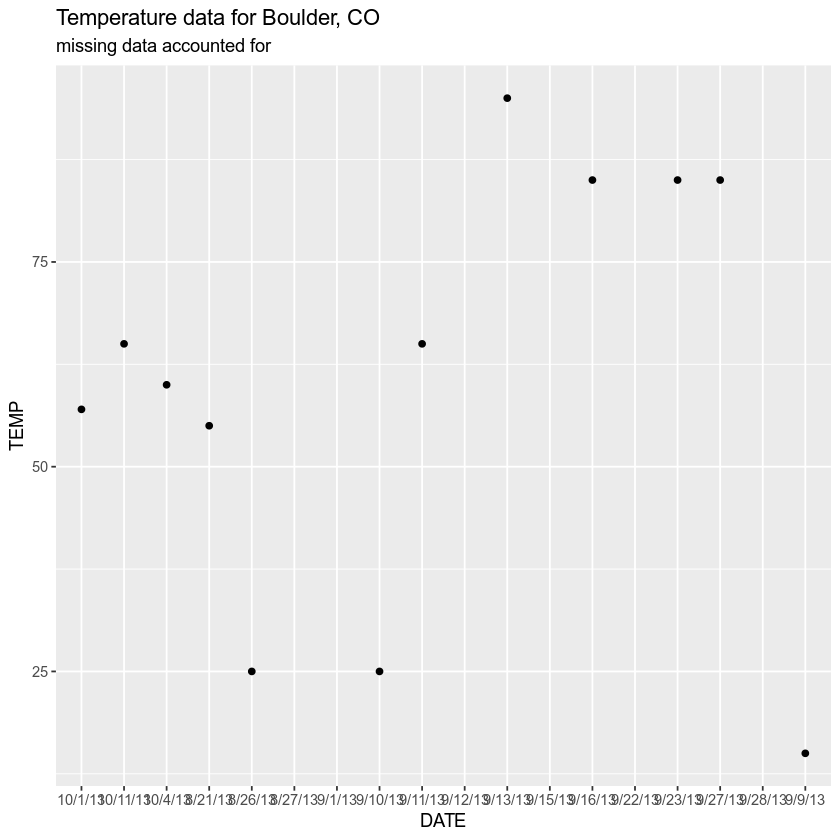
Optional Challenge
- Question: Why, in the the example above did hateful(boulder_precip$avg_temp) render a value of NA?
- Question: Why, in the the example in a higher place did mean(boulder_precip$avg_temp, na.rm = TRUE) also render a value of NA?
# Extract those elements which are not missing values. heights [ ! is.na ( heights )] ## [one] 2 four 4 6 # Returns the object with incomplete cases removed. The returned object is diminutive. na.omit ( heights ) ## [1] two 4 4 6 ## attr(,"na.action") ## [1] four ## attr(,"class") ## [one] "omit" # Extract those elements which are complete cases. heights [ complete.cases ( heights )] ## [one] 2 4 4 half-dozen Challenge Activity
- Question: Why does the following piece of code return a alarm?
sample <- c ( ii , 4 , 4 , "NA" , 6 ) mean ( sample , na.rm = True ) ## Alert in mean.default(sample, na.rm = TRUE): statement is not numeric or ## logical: returning NA ## [1] NA - Question: Why does the warning message say the statement is not numeric?
Source: https://www.earthdatascience.org/courses/earth-analytics/time-series-data/missing-data-in-r-na/
Post a Comment for "Read Table in R With Missing Values"